Bioinformatics for understanding cells, organisms, and the biosphere
Bioinformatics
Bioinformatics, also called computational biology, is a relatively new discipline that has emerged as the result of high-throughput experimental technologies in genome research and post-genome research. The data generated by these technologies include genome sequence data, cDNA (mRNA) sequence data, gene expression data by DNA chips or microarrays, genome variation data such as SNPs, and 3D coordinates of protein structures. The increasing amount of such genomic information is the basis for understanding principles of how higher-order biological systems, such as cells, organisms, and the biosphere, are formed, as well as for medical, industrial, and other practical applications. However, current bioinformatics technologies cannot readily uncover higher-level complexity of such biological systems, although they are quite effective for finding and characterizing building blocks of genes and proteins. We at the Bioinformatics Center of the Institute for Chemical Research, Kyoto University develop knowledge-based methods for uncovering higher-order systemic behaviors of cells and organisms from genomic information. The reference knowledge is stored in KEGG, Kyoto Encyclopedia of Genes and Genomes, and associated bioinformatics technologies are developed for both basic research and practical applications.
New concept of database
Large amounts and different types of biological data are stored in various databases worldwide. For example, the Entrez System of the U.S. National Center for Biotechnology Information integrates PubMed for biomedical literature, GenBank/RefSeq for nucleic acid and protein sequences, PubChem for small chemical compounds, and many more databases. Here the database is a repository of all published data in a given domain, and the integrated system such as Entrez forms an information infrastructure for biomedical sciences. However useful such a resource may be for searching and retrieving individual data, it does not provide an overall picture of how the biological system works. We believe that an ultimate goal of bioinformatics is a complete computer representation of cells and organisms, which will enable computational prediction of higher-level complexity such as cellular processes and organism behaviors. Here the database is not a simple repository; it is a computer representation of the biological system. Developing this type of database is like synthesizing a virtual cell or an organism from the building block information currently available. Searching against such a database is like doing an in silico experiment on cells or organisms. KEGG is a practical implementation of this database concept.
The KEGG resource
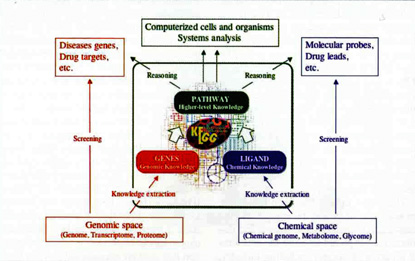
The KEGG database project was initiated in our laboratory in 1995, the last year of the first five-year phase of theJapanese Human Genome Program. It, continued in the second five-year phase, and was significantly expanded under the Millennium Project. Figure 1 illustrates an overall architecture of KEGG, where genomic information (GENES database) and chemical information (LIGAND database) are integrated in terms of network information (PATHWAY database). In contrast to traditional bioinformatics technologies for screening of useful building blocks (molecules), our approach is first to understand wiring diagrams (molecular interaction networks) of building blocks and then to find functions and utilities of biological systems as a whole. KEGG is a reference knowledge base containing current knowledge on such wiring diagrams, and it is used worldwide as a unique resource for reconstructing metabolic and other cellular processes from genomic information and for understanding systemic functional meanings and utilities (Table 1).
| Database service | Number of links* |
|---|---|
| NCBI (External Link) | 29,800 |
| ExPASy (SwissProt) (External Link) | 18,300 |
| EBI (External Link) | 13,200 |
| GenomeNet (KEGG) (External Link) | 9,430 |
| DDBJ (External Link) | 620 |
Table 1. Major biological database services.
* The number of pages linked to each database service site according to the Google links search on July 16, 2005.
Integration of Genomics and Chemistry
In the fall of 2003, the U.S. National Institutes of Health announced the Roadmap, which contained new chemical genomics initiatives for screening of useful chemical compounds such as imaging probes and drug leads. While traditional genomics and post-genomics have contributed to our knowledge on the genomic space of possible genes and pro-biological system, chemical genomics will give us a glimpse of the chemical space of possible compounds and reactions that exist as an interface between the biological system and the natural environment. The wiring diagram information in KEGG can then be extended to include both endogenous and exogenous molecules, which would enable reconstruction of the still higher biological system, the biosphere. A joint venture between the Bioinformatics Center and the School of Pharmaceutical Sciences in Kyoto University has been undertaken as a 21st Century Center of Excellence (COE) program, aiming at developing new bioinformatics technologies integrating genomics and chemistry for the purpose of pharmaceutical and medical applications.

A distance learning linkup with a laboratory in Tokyo

A cluster of database servers supporting the core operations of KEGG

Minoru Kanehisa
Born in 1948.
Specialized Research Field: Bioinformatics
Graduate of the doctoral program, Graduate School of Science, The University of Tokyo D.Sc., The University of Tokyo Professor and Director, Bioinformatics Center, Institute for Chemical Research, Kyoto University
URL : Kanehisa Laboratory (External Link)
Overseas, they don't call me "Professor Kanehisa"; they call me "Mr. KEGG"
Currently, GenomeNet is accessed about 10 million times per month. Professor Kanehisa set up the GenomeNet service in 1991. The international project to decipher the human genome had only just begun, and the Internet had yet to spring into existence. In 1995, he started using his own database that was to form the core of the project, KEGG. Since then, KEGG has become a unique database, winning international recognition and becoming widely used. However, Prof. Kanehisa has described the structure of the project as "feeling a bit like creating life inside a computer." Prof. Kanehisa, who originally majored in physics, became a pioneer in the field of bioinformatics in Japan through his research at the Los Alamos National Laboratory in America. From his initial use of models to view the world, Prof. Kanehisa's approach underwent a 180 degree change, to using data to explore the world. This change was, simply put, brought on by exposure to a different culture. Based on his experiences, Prof. Kanehisa remains a strong proponent of cooperation among "cultures". While sharing results and conducting joint research in collaboration with industry, particular emphasis is being placed on the human resource development of people knowledgeable in the field of bioinformatics. In cooperation with Kyoto City, "Sakagura VIL" was set up in a converted sake brewery, in order to support the development of bioinformatics venture businesses. Since bioinformatics is such a new field, it has attracted a range of people with diverse backgrounds. New ideas springing from the merging of disparate cultures this is one of the most important sources of energy driving the vigorous research of Prof. Kanehisa and the Kanehisa Laboratory