Research reveals how cells achieve self-organization
Researchers from Kyoto University have developed a simple model for cell motion, which may explain how large groups of cells are able to coordinate and regulate their motion in tissues.
The complex motion of cells is crucial for the proper function of biological systems. For example, when an embryo is forming from a fertilized egg, its cells have to coordinate their movement to ensure healthy growth. In tumors, this regulation of motion breaks down, allowing cancerous cells to run amok.
Developing an understanding of how cells move and interact could prove extremely valuable, and has attracted the attention of researchers in biology, medicine, and physics.
Previous work has shown that cells respond to their environment through complex bio-mechanical processes. For instance, cells repel each other when they come into contact, a process called 'contact inhibition of locomotion'. This is believed to be important for the self-organization of cells.
In a paper published in Scientific Reports , Ryoichi Yamamoto and his team at Kyoto University were able to build a simple model that reproduces many of these complex cell behaviors.
The model represents cells as two disks connected by a spring, one for the pseudopod --the front of the cell -- and the other for the cell body. By coupling the pseudopod's movement to the extension of the spring, the team found that cells naturally exhibit contact inhibition of locomotion; they slow down and change direction upon contact, and stop moving at sufficiently high cell densities.
Further study revealed that the shape of the cell greatly influences collective motion. Those with a large front quickly align their motion, suggesting that cells might have evolved broader fronts to facilitate alignment.
The researchers were surprised to observe that the cells exhibited a dynamic state corresponding to what we know as traffic jams: density waves traveling against the cells' direction of motion.
This allowed the researchers to not only draw a connection to the traffic flow of cells, but to also provide a possible explanation for previous observations of backwards-travelling waves in cell colonies.
This work highlights how simple mechanisms can give rise to complex behaviors in large systems.
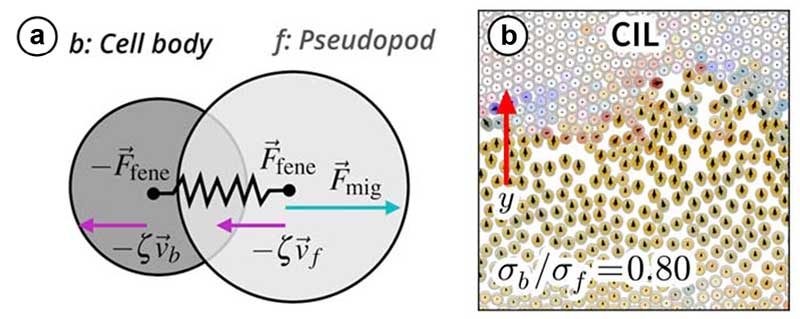
(a) Illustration of the cell model incorporating contact inhibition of locomotion: a cell consists of two disks connected by a spring. The motility is proportional to the cell extension (distance between disks).
(b) Simulation snapshot of cells running into a traffic jam
Paper Information
【DOI】 https://doi.org/10.1038/s41598-017-05321-0
【KURENAI ACCESS URL】 http://hdl.handle.net/2433/226397
Simon K. Schnyder, John J. Molina, Yuki Tanaka & Ryoichi Yamamoto (2017). Collective motion of cells crawling on a substrate: roles of cell shape and contact inhibition. Scientific Reports, 7, 5163.





